Machine Learning
Background:
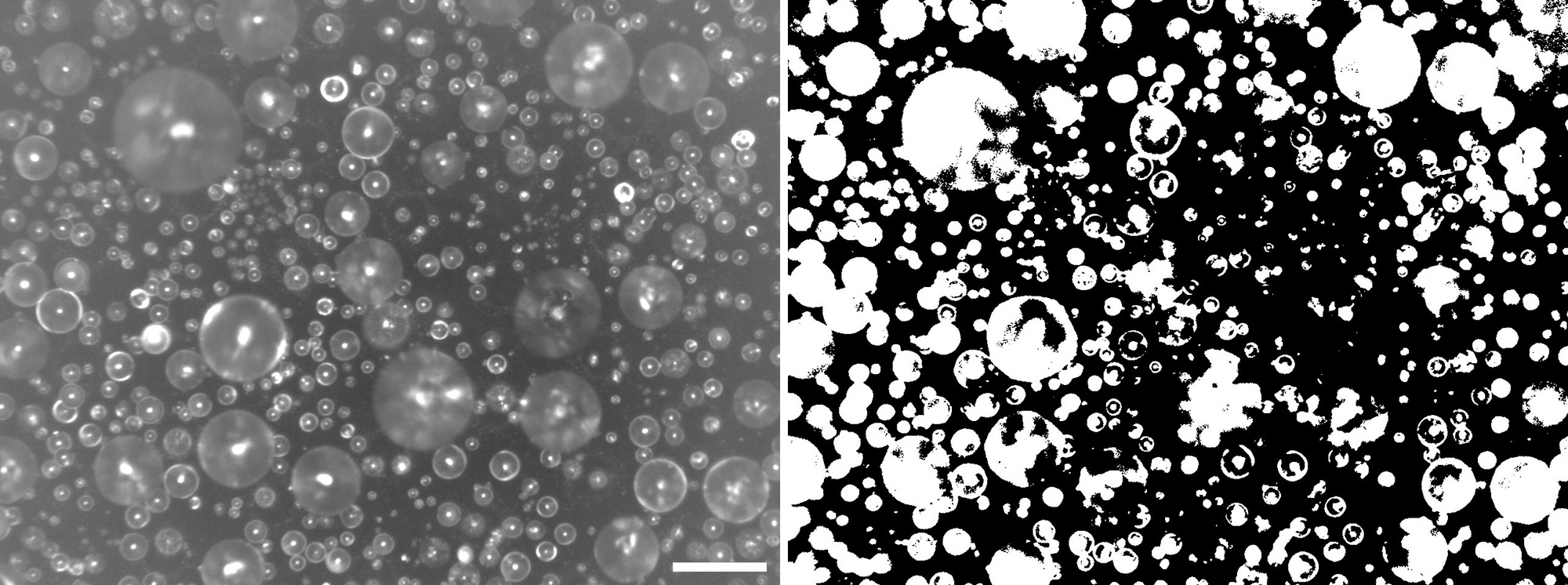
I need an efficient and accurate method to annotate the droplets contained in the Time-lapse images. It is challenging for the "traditional image analysis" methods, such as image thresholding (Figure 1). Because of few things:
- Various focus between droplets
- Various intensity between regions
- Segmentation difficulties
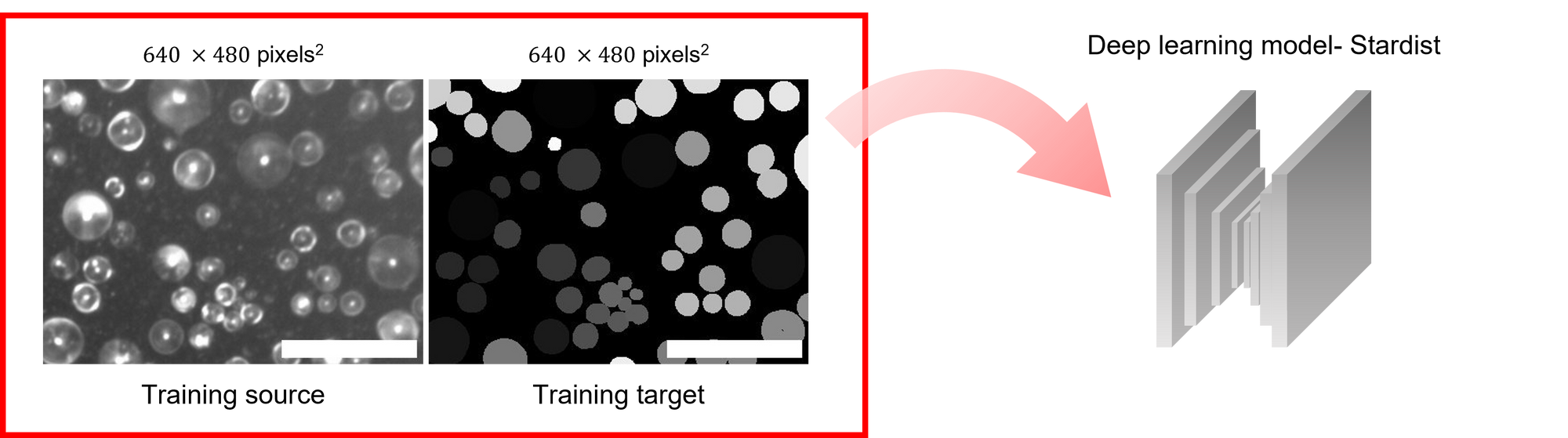
To overcome the challenge, I adopted the machine learning approach. I found a open-source model named Stardist, which is aimed to detect the Star-shaped polygon1. I only need to prepared the training sets to get the model (Figure 2).
Challenge:
Because my image is composed of the droplet at various size, the majority is ~10-6 m and there are a few droplet at 10-4 m. In consequence, the training volume of the "big droplet" was less than the "small droplet", led to the inaccurate model if I did not enhance the training volume of "big droplet". However, it will take me tremendous effort to prepared the enough training volume for the "big droplet".
Solution:
Here I would like to share a little trick. First, I prepared a few training set and trained the model. Second, I used the trained-model to annotated the "small droplet". Therefore, I only need to annotate the "big droplet" to prepare the good enough training sets.
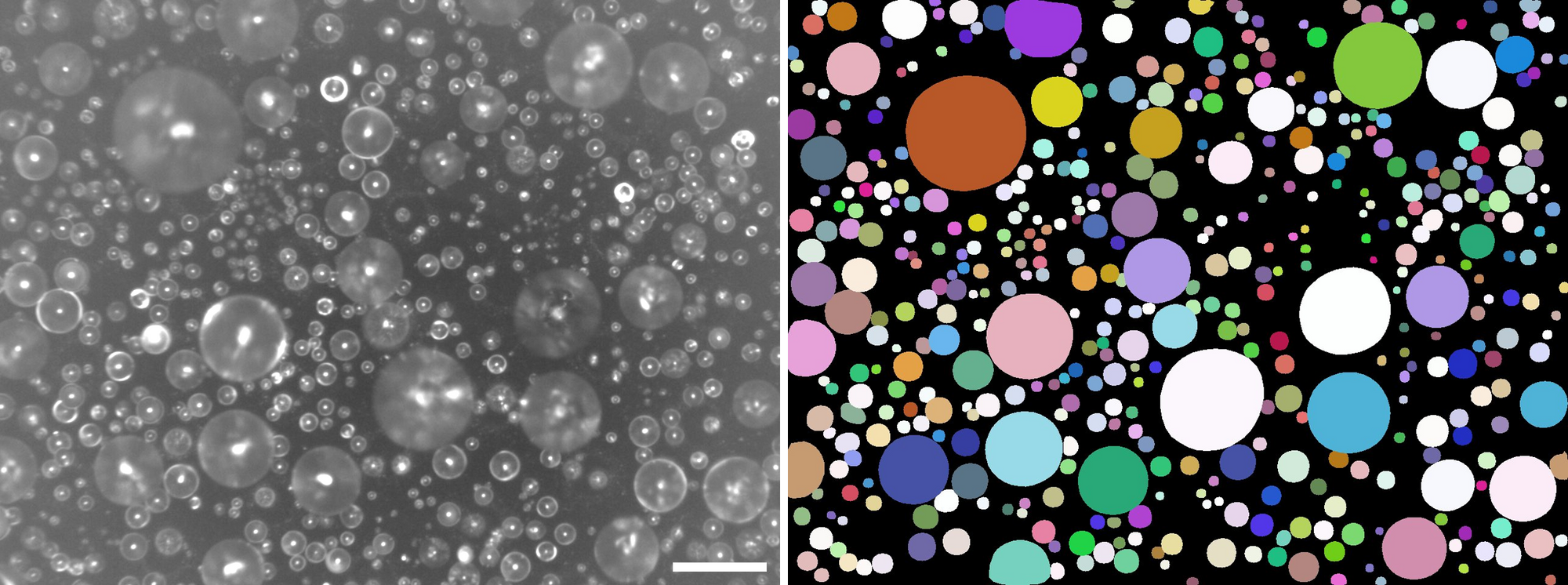
The result is satisfying (Figure 3)!
Reference:
- Schmidt, U., Weigert, M., Broaddus, C., & Myers, G. (2018). Cell detection with star-convex polygons. In Medical Image Computing and Computer Assisted Intervention–MICCAI 2018: 21st International Conference, Granada, Spain, September 16-20, 2018, Proceedings, Part II 11 (pp. 265-273). Springer International Publishing.